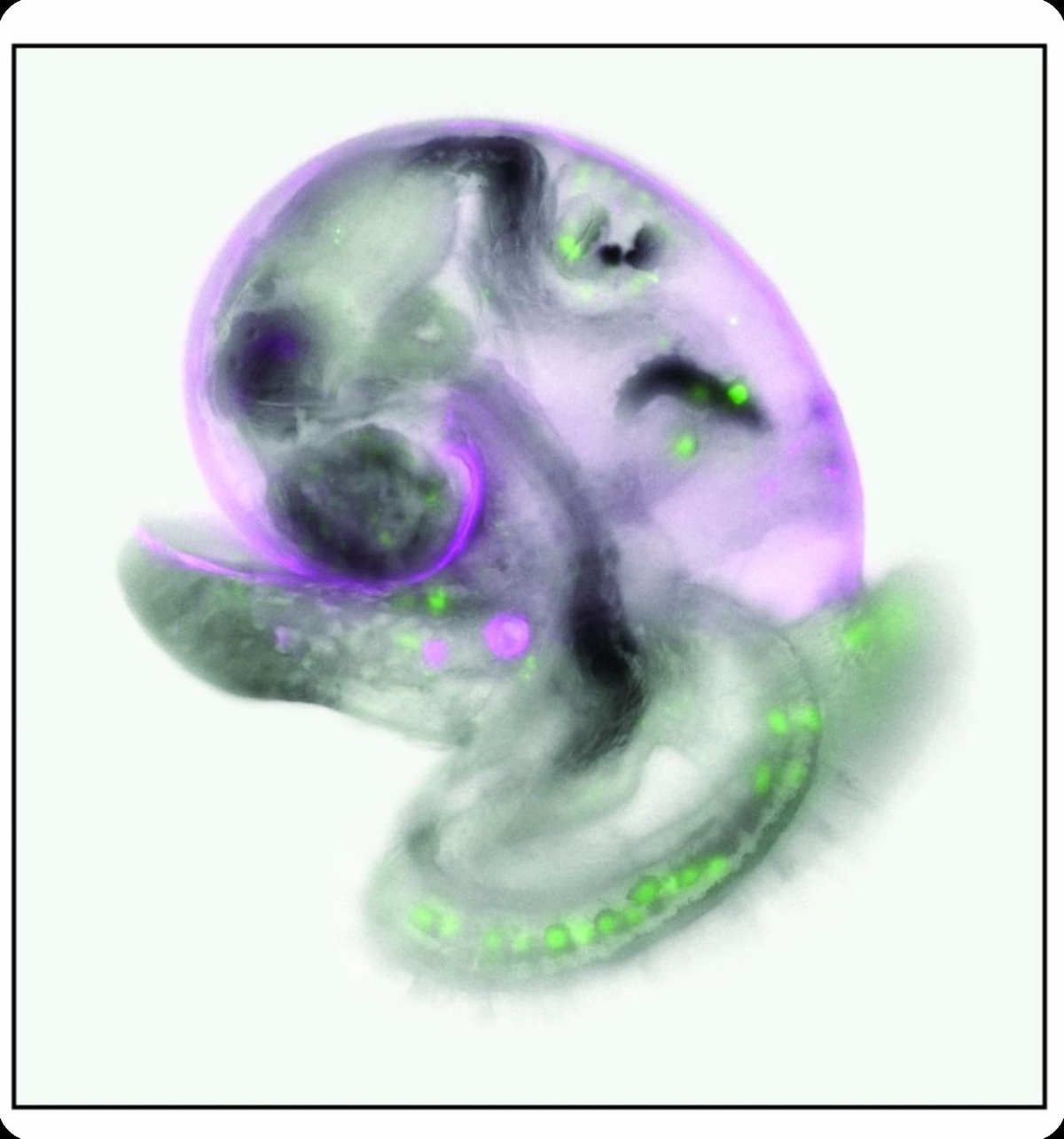
Modelling ecoevolutionary dynamics

This involves two major strands: Firstly, the development of stochastic individual based models to understand the evolutionary responses to environmental change, especially in relation to dispersal and range shifts, and; secondly the application of sophisticated quantitative genetic analyses on long-term, large scale multigenerational datasets to test key hypotheses around evolution and coevolution of life history traits, mating systems and fitness.
Functional and ecological genomics

This research is directed to providing insight into the evolutionary adaptations of organisms to environmental change, and also addresses key challenges in agriculture and aquaculture by facilitating selective breeding programmes.
- Alex Douglas
- Cecile Gubry-Rangin
- Cath Jones
- Kara Layton
- Fabio Manfredini
- Sam Martin
- Gareth Norton
- Stuart Piertney
- Adam Price
- Vicky Sleight
- John Speakman
- Tom Vogwill
Molecular Ecology
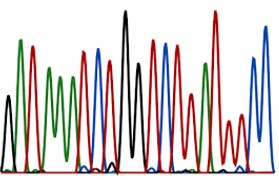
There has also been considerable emphasis on understanding the causes and consequences of variation in levels of neutral and adaptive genetic diversity among individuals and populations - how do population dynamics, demographic history and environmental change influence extant genetic variation and how does this impinge on individual fitness, behaviour,population viability and the evolution of life histories.
Molecular data is also being utilised for resolving evolutionary relationships among species and for identifying the processes involved in shaping their distribution
This work involves neutral polymorphisms such as SNPs and microsatellites, ecologically meaningful candidate loci such as the MHC, and latterly genome-wide variation gleaned from next-generation sequencing.
- Cecile Gubry-Rangin
- Cathy Jones
- Lesley Lancaster
- Kara Layton
- Fabio Manfredini
- Stuart Piertne y
- Vicky Sleight
- Tom Vogwill
Comparative genomics
Many species underwent evolutionary adaptations that allowed them to thrive in their environment. These adaptations can include changes to anatomy, physiology and metabolism and are driven by modification to genes over several generations. By comparing genomes of different species, the defining characteristics or similarity can be identified. This research is directed to provide insight into the cellular functioning of species, how this may have changed through evolution and what the impact may be for their conservation.
Eco-Evo-Devo