
B.Sc. (Tokyo University of Science, 2003), PhD (Tokyo University of Science, 2008)
Lecturer
- About
-
- Email Address
- t.kubota@abdn.ac.uk
- Telephone Number
- +44 (0)1224 437494
- Office Address
Room 2.19
Institute of Medical Sciences
University of Aberdeen
Foresterhill
Aberdeen
AB25 2ZD- School/Department
- School of Medicine, Medical Sciences and Nutrition
Biography
Takashi Kubota studied Molecular Biology and did his PhD at the Tokyo University of Science. Takashi then moved as a postdoctoral fellow to the University of Aberdeen in 2008 and has worked with Prof. Anne Donaldson until 2014.
Takashi establishes his research group as an MRC Career Development Fellow in the Institute of Medical Sciences at the University of Aberdeen. Since 2019, he is a Lecturer in the Institute of Medical Sciences.
Qualifications
- BSc Molecular Biology2003 - Tokyo University of Science
- MSc Molecular Biology2005 - Tokyo University of Science
- PhD Molecular Biology2008 - Tokyo University of Science
Internal Memberships
Co-organiser of the PI seminar series joint between the IMS and the Rowett institute (2016-2017)
Organiser of the IMS PI seminar series (2017-)
- Research
-
Research Overview
I am interested in mechanisms of drug resistance and, particularly would like to know how fungi develop resistance to antifungal drugs.
Fungi infect billions of people around the world each year, and Candida species are the second most numerous agents of invasive fungal infections. The global incidence of life-threatening Candida infections stands at ~400,000 cases annually, and the emergence of multidrug-resistance strains of Candida species is a major global concern.
Our group is investigating the mechanism of drug resistance caused by over-expression of multidrug transporter genes (e.g., PDR5 in Saccharomyces cerevisiae and CDR1 in Candida species). We recently discovered that the chromatin remodeller SWI/SNF and the histone chaperone Rtt106 drive expression of multidrug transporter genes, conferring drug resistance in Candida glabrata and its close relative S. cerevisiae (Nikolov et al. 2022 Nat Commun).
We are actively using a novel method to identify proteins assembled on promoters (Nikolov et al. 2022 Nat Commun), and successfully improved this method recently (see Figure below). Using this method, we are now investigating dynamic regulation of gene expression.
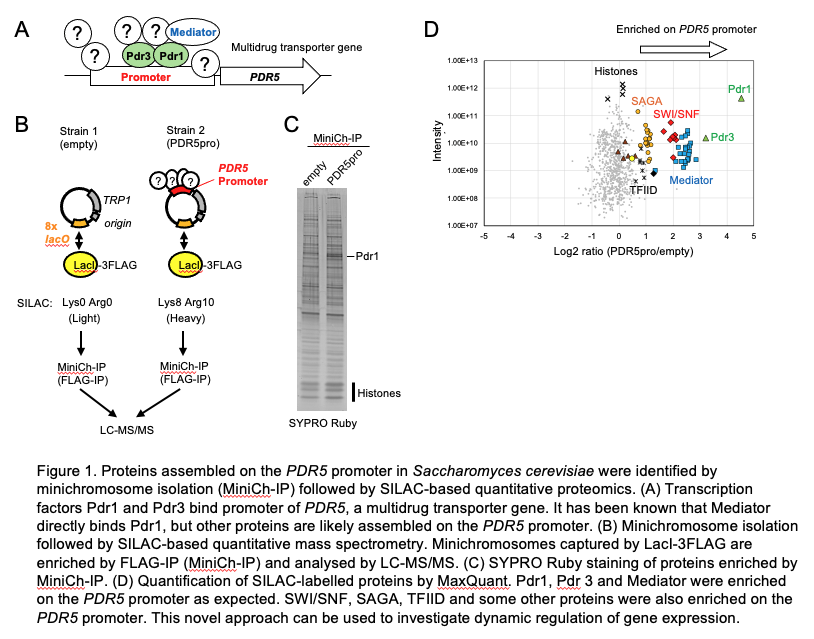
We are also investigating how the protein level of multidrug transporters is regulated, and whether we can manipulate it to tackle drug resistance.
Research Areas
Accepting PhDs
I am currently accepting PhDs in Biomedical Sciences.
Please get in touch if you would like to discuss your research ideas further.
Research Specialisms
- Genetics
- Molecular Biology
- Microbiology
Our research specialisms are based on the Higher Education Classification of Subjects (HECoS) which is HESA open data, published under the Creative Commons Attribution 4.0 International licence.
Current Research
How is expression of multidrug transporter genes regulated in Candida species?
Our group is currently investigating mechanisms regulating expression of multidrug transporters and drug resistance in the pathogenic fungi Candida species. We are particularly interested in transcriptional regulation of multidrug transporter genes mediated by histone chaperones and chromatin remodellers. We developed a technique to identify proteins assembled on promoters in a model organism S. cerevisiae and are going to apply this technique to Candida species.
We have recently improved this method for S. cerevisiae (see Figure shown above), and are investigating dynamic regulation of gene expression.
How is the protein level of multidrug transporters regulated in yeasts?
We are investigating how the level of multidrug transporter proteins regulated in yeasts, particularly, their degradation mechanism. We are also developing a new strategy to decrease their protein level to tackle drug resistance.
Past Research
How do the RFC family members (Elg1, Ctf18 and Rad24) maintain genome integrity?
PCNA is a ring-shaped homotrimeric complex, which clamps DNA polymerase to DNA during replication and acts as a sliding scaffold for many replication and repair proteins. RFC loads PCNA on DNA. We discovered that the Elg1 RFC-like complex (Elg1-RLC) unloads PCNA (Kubota et al. 2013 Molecular Cell 50: 273; Kubota et al. 2013 Cell Cycle 12: 2570). Our group further found that Elg1-RLC unloads PCNA from DNA gnome-wide and after Okazaki fragmant ligation (Kubota et al. 2015 Cell Reports 12: 774). Elg1 is critical for genome maintenance, and we found that PCNA retention on DNA into G2/M phase is the main cuase of genome instability observed in yeast cells lacking Elg1 (Johnson et al. 2016 Cell Reports 16: 684). Recently, we found that PCNA retention controlled by the Elg1 complex is critical for efficient mismatach repair (Paul Solomon Devakumar et al. 2019 Nucleic Acids Research). In this recent study, we also isolated 'retention-prone' and dissociation-prone' PCNA mutants, which will be useful for further investigation of PCNA-related events.
We found that Ctf18-RLC, which can load and unload PCNA in vitro, is important for activation of S-phase checkpoint (Kubota et al. 2011 Mol Cell Proteomics) and cohesion establishments.
Supervision
My current supervision areas are: Biomedical Sciences.
Teera Leepattarakit (PhD student: 2023-present)
Vladislav Nikolov (PhD student: 2016-2021)
Lovely Jael Paul Solomon Devakumar (PhD student: 2015-2018)
Research Assistant/Postdoc:
Dr Elizabeth Hughes (2024- present)
Dr Catherine Johnson (2014-2018, 2022)
Funding and Grants
2024- BBSRC New Investigator Grant
2023- Tenovus Scotland Small Pilot Grant
2022- Early Career Researcher Grant - Medical Research Scotland
2021- The Carnegie Trust Research Incentive Grant
2014 - 2019 MRC Career Development Fellowship
2013 BBSRC Researcher Co-Investigator (project grant with Prof Anne Donaldson)
- Teaching
-
- Publications
-
Page 1 of 2 Results 1 to 10 of 15
Use of Nuclear and Chromatin Enrichment Procedures for Quantitation of Yeast DNA Replication Proteins Using SILAC
SILAC: Methods and Protocols. Luque-Garcia, J. L. (ed.). 1 edition. Springer US, pp. 209-218, 10 pagesChapters in Books, Reports and Conference Proceedings: Chapters- [ONLINE] DOI: https://doi.org/10.1007/978-1-0716-2863-8_17
SWI/SNF and the histone chaperone Rtt106 drive expression of the Pleiotropic Drug Resistance network genes
Nature Communications, vol. 13, 1968Contributions to Journals: ArticlesLigation of newly replicated DNA controls the timing of DNA mismatch repair
Current Biology, vol. 31, no. 6, pp. 1268-1276.E6Contributions to Journals: ArticlesEffective mismatch repair depends on timely control of PCNA retention on DNA by the Elg1 complex
Nucleic Acids Research, vol. 47, no. 13, pp. 6826-6841Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1093/nar/gkz441
- [OPEN ACCESS] http://aura.abdn.ac.uk/bitstream/2164/12637/1/gkz441.pdf
- [ONLINE] View publication in Mendeley
Identification of Elg1 interaction partners and effects on post-replication chromatin re-formation
PLoS Genetics, vol. 14, no. 11, e1007783Contributions to Journals: ArticlesPCNA Retention on DNA into G2/M Phase Causes Genome Instability in Cells Lacking Elg1
Cell Reports, vol. 16, no. 3, pp. 684-695Contributions to Journals: ArticlesReplication-Coupled PCNA Unloading by the Elg1 Complex Occurs Genome-wide and Requires Okazaki Fragment Ligation
Cell Reports, vol. 12, no. 5, pp. 774-787Contributions to Journals: ArticlesDefinition of the transcription factor TdIF1 consensus-binding sequence through genomewide mapping of its binding sites
Genes to Cells, vol. 20, no. 3, pp. 242-254Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.1111/gtc.12216
Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex
Genes & Development, vol. 28, no. 4, pp. 372-383Contributions to Journals: ArticlesIs PCNA unloading the central function of the Elg1/ATAD5 replication factor C-like complex?
Cell Cycle, vol. 12, no. 16, pp. 2570-2579Contributions to Journals: Articles- [ONLINE] DOI: https://doi.org/10.4161/cc.25626
- [OPEN ACCESS] http://aura.abdn.ac.uk/bitstream/2164/3397/1/Cell_Cycle.pdf
